FAYETTEVILLE, Ark. — A plant biologist at the University of Arkansas will use a nearly $440,000 grant from the National Science Foundation to reconstruct the origin and evolution of the genetic makeup of diatoms, one of the world’s most diverse groups of microalgae.
“Diatoms are microscopic, single-celled algae that are found in oceans, lakes and rivers; practically anywhere where there is sunlight and moisture,” said Andrew J. Alverson, an assistant professor of biological sciences in the J. William Fulbright College of Arts and Sciences. “They are prolific photosynthesizers. They produce the oxygen for one of every five human breaths, so they’re extremely important in global cycling of carbon and oxygen.”
With an estimated 100,000 species, diatoms represent one of the most diverse lineages in the “tree of life.” Only a handful of diatoms have had their genome sequenced, however. This project will double the number of diatoms with fully sequenced genomes.
Under the grant, Alverson will examine genomic factors that will help explain the extraordinary diversity of diatoms. His research team will sequence the full genomes of five species and all the expressed genes – the parts of the genome that encode for proteins – from another 200 species. Using resources available through the Arkansas High Performance Computing Center at the University of Arkansas, Alverson’s team will use mathematical and statistical models to reconstruct the historical pattern of large-scale genomic changes.
One such genomic change results from horizontal gene transfer, which is the transfer of genetic material between entirely different species.
“When researchers sequenced the first diatom genomes, they found that the genomes contained hundreds of genes that appear to have been acquired by horizontal gene transfer from bacteria,” Alverson said. “This is gene transfer across kingdoms, across species that last shared a common ancestor several billion years ago. These genes appear to encode for several diverse and important functions that may have played important roles in the diversity of diatoms, including the conditions they can survive in and their physiology.”
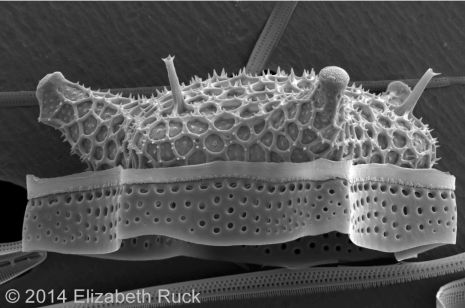
In addition to their importance ecologically, diatoms are also notable for their cell wall. It is made out of silica, which is essentially glass.
“They are one of a very few organisms that make their cell wall this way,” Alverson said. “Scientists have marveled at their ornate glass walls since the late 18th century. They produce these cell walls on the nano scale, replicating their intricate patterns with really high fidelity. There is a lot of interest in finding out how they do this.”
The NSF grant will fund collaboration between Alverson’s research laboratory and the lab led by Norman Wickett, assistant conservation scientist in genomics and bioinformatics at the Chicago Botanic Garden. The research will be showcased through an exhibit at the botanic garden that will highlight the numerous parallels between diatoms and flowering plants.
Alverson is a member of several professional societies and is an associate editor for two scientific journals, Diatom Research and Phycologia. He came to the University of Arkansas in January 2012 from Indiana University, where he studied plant genomics under a Ruth L. Kirschstein Postdoctoral Research Fellowship from the National Institutes of Health.
Contacts
Andrew J. Alverson, assistant professor biological sciences
J. William Fulbright College of Arts and Science
479-575-7975, aja@uark.edu
Chris Branam, research communications writer/editor
University Relations
479-575-4737,
cwbranam@uark.edu